- Home
- Genomes
- Genome Browser
- Tools
- Mirrors
- Downloads
- My Data
- Projects
- Help
- About Us
Genotype Tissue Expression (GTEx) Project Resources at UCSC
The Genotype Tissue Expression (GTEx) Project is a data resource and tissue bank established by the National Institutes of Health Common Fund, in order to study the relationship between genetic variation and gene expression in a variety of human tissues. A total of 53 tissues from nearly 1000 individuals have been studied by genomic and RNA sequencing. These resources are valuable for exploring the genetic basis of human diseases. Extensive data access and visualizations are available from the GTEx Portal, a resource provided by the GTEx Laboratory, Data Analysis and Coordinating Center at the Broad Institute.
The UCSC Genome Browser provides visualization resources for selected summary data from the GTEx project, including these tracks and hubs:
- GTEx Gene track
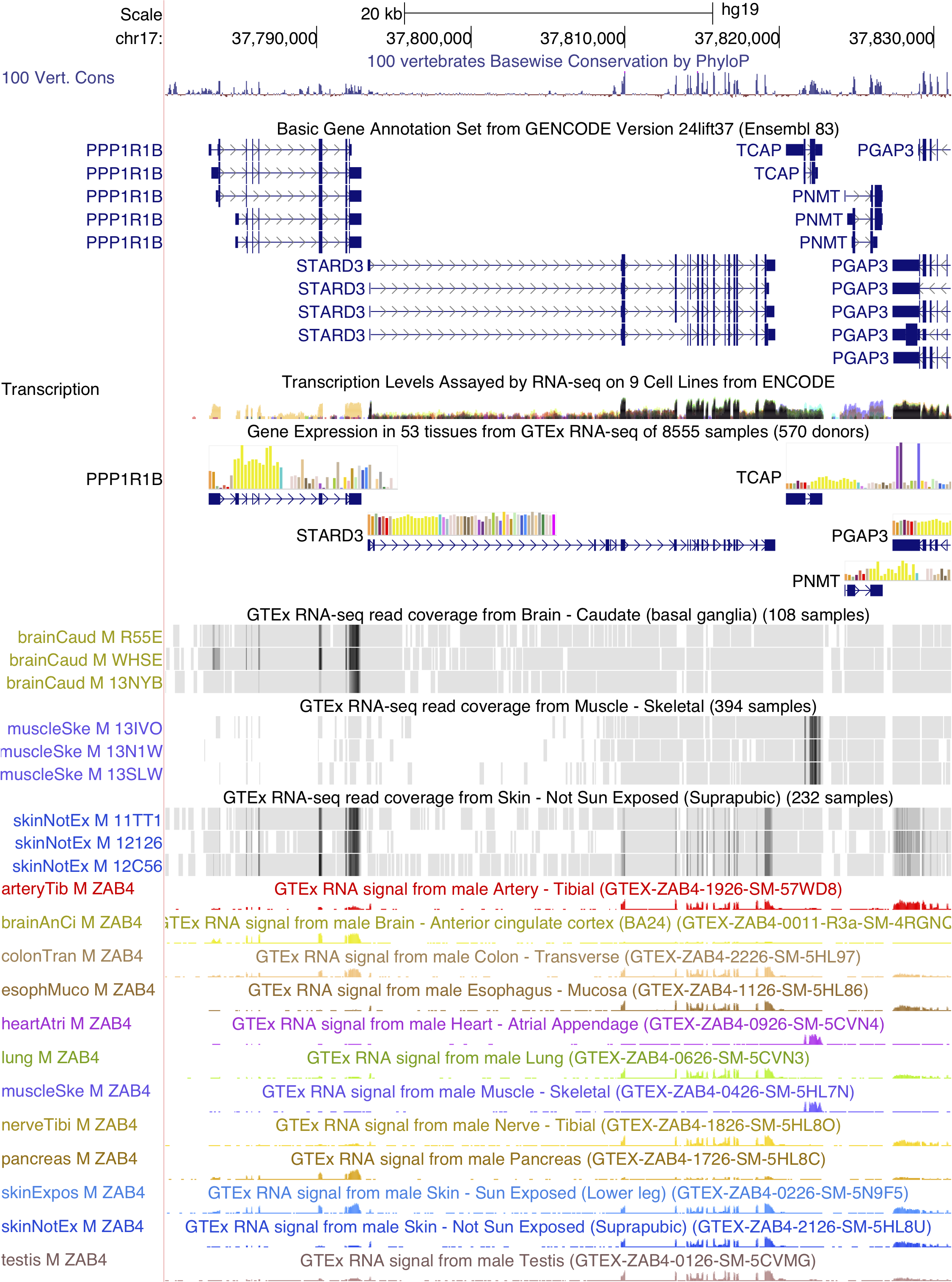
- Gene Expression in 53 tissues from GTEx RNA-seq of 8555 samples (570 donors)
- GTEx Transcript track
- Transcript Expression in 53 tissues from GTEx RNA-seq of 8555 samples (570 donors)
- GTEx Combined eQTL track
- Combined Expression QTLs in 44 Tissues from GTEx
- GTEx Tissue eQTL track
- Expression QTLs in 44 tissues from GTEx
- GTEx Signal hub
- GTEx RNA-seq of 53 tissues (570 donors)
- GTEx Analysis hub
- Allele-Specific Expression in 53 Tissues from GTEx Analysis
- Distant variants affecting gene expression (trans-eQTLs) from GTEx Analysis
- GTEx V8 Gene track
- Gene Expression in 54 tissues from GTEx RNA-seq of 17,382 samples (948 donors)
GTEx Tissue Body Map
This interactive graphic identifies the tissues sampled by GTEx for gene expression studies. Some GTEx browser tracks include the Body Map in the track configuration page, to facilitate limiting track display to tissues of interest. A stand-alone page with the interactive graphic is here.
Credit: jwestdesign
GTEx in the browser
Below is a screenshot of a public session that highlights the GTEx gene expression track and GTEx RNA-seq signal hub for the GTEx Resources in the Genome Browser blog article. The session shows a genomic region where 5 genes exhibit different patterns of tissue-specific expression.