- Home
- Genomes
- Genome Browser
- Tools
- Mirrors
- Downloads
- My Data
- Projects
- Help
- About Us


Genome Browser
We offer a REST API available under the menu (Downloads then REST API) that can return almost all data available in the Browser in JSON format. It also has additional endpoints, such as /list/schema, that can retrieve the internal organization of track data stanzas (trackDb), which is otherwise unavailable.
Feel free to contact us if you are interested in attending a workshop, or meeting someone from the team to collaborate, get help, or ask any questions at the meetings. See our Poster Gallery.

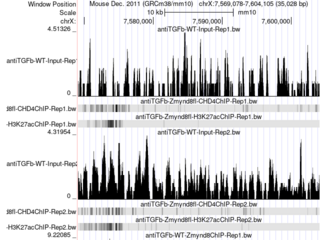
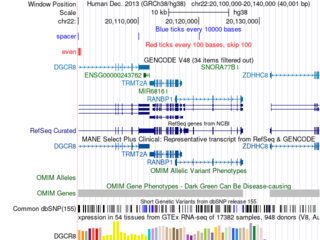
Click the images to explore the latest Public Sessions on our main site.